COVID-19 Multiomics Research: Strategies, Insights and Tools

Complete the form below to unlock access to ALL audio articles.
The integration of multiple "omics" approaches, known as multiomics or multi-omics, has become a major focus in biomedical research, as researchers seek ways to understand biological mechanisms in greater depth. While omics fields such as genomics, transcriptomics, proteomics and metabolomics provide information about DNA, RNA transcripts, proteins and metabolites, respectively, multiomic studies allow multiple biomolecule types to be examined together and offer the potential to obtain a more holistic, pathway-oriented view of biology.
Having the potential to generate, share and mine important biological information at speed is valuable in any research setting. In the context of the current pandemic, however, there is a heightened sense of urgency, and researchers are seeking ways to unravel the molecular basis of COVID-19. Multiomics approaches are being used in COVID-19-related studies of pathophysiology, biomarkers, immune responses, disease severity prediction and beyond.
In practice, multiomic datasets are large, contain information derived using multiple technologies and feature many potentially confounding variables. Therefore, the management and exploration of multiomic datasets is challenging. In this article, we explore multiomic approaches to COVID-19 research and clinical omics and explore some of the computational and statistical tools being developed to support multiomic analyses.
Novel Virus Detection Using a Comprehensive Viral Research Panel

Infections from novel viral species and strains present a serious and recurring threat to global public health. Responding to these threats critically depends on the ability to identify and develop tests for the viral agent, which is a significant challenge given the diverse and rapidly-evolving sequence space of human viruses. Download this app note to discover a novel virus discovery and surveillance approach that provides high sensitivity detection of known viral species and delivers enrichment and complete characterization of novel viral sequences.
View App NoteSponsored Content
Aims and strategies of COVID-19 multiomics studies
How can multiomics approaches help us pave a way out of the COVID-19 pandemic? As explained by Al J Abadi, genomics data scientist at Melbourne Integrative Genomics, University of Melbourne, it’s all about tracing molecular footprints: “Put simply, the SARS-CoV-2 virus, like any other virus, triggers a host immune response and leaves a footprint in patients' proteome as well as the metabolome. This footprint varies among different patients – some of whom also experience a more severe form of the disease. Characterizing the variability in these omics alterations and their interactome is only possible through multiomics approaches and could help gain a better understanding of the disease mechanism and how we can combat it more efficiently.”
Evgenia Shishkova, assistant staff scientist at the Coon Laboratory, University of Wisconsin-Madison, explains that the aims of multiomics studies are very project-dependent: “Whether your goal is to create a resource, or you have a more targeted approach, the technology lets you do either. But more often than not, multiomic datasets end up being really big. So even if you do have a targeted question in mind, you end up having this massive resource that others could benefit from.”
Not only do multiomics studies vary in their size, underlying goals and types of data used, but also their approaches to data integration. Algorithms used to integrate omics data can be categorized as supervised, semi-supervised, or unsupervised, and the integration can occur at different stages of analysis.1 Within these categories, there are many different analytical subtypes, such as matrix factorization, Bayesian, network-based and multiple-step approaches which use different modeling and statistical techniques to identify data patterns.
Interrogate the SARS-CoV-2 Genome To Get More Answers

Studying the SARS-CoV-2 genome can provide valuable insights into its evolution and transmission, as well as aiding surveillance and vaccine development efforts to help control the COVID-19 pandemic. Download this app note to discover an RNA sequencing approach that can accurately identify low copy number viral pathogens and is sensitive enough for use on low concentration, low quality and degraded samples.
Download App NoteSponsored Content
Building a COVID-19 knowledge base using multiomics approaches
Multiomic data serve as a resource to be explored by the wider scientific community and provide critical supporting evidence of the key biological processes modulated in people with COVID-19. Physiological changes associated with COVID-19 could be identified on a molecular level, such as those related to lipid transport, complement system activation, vessel damage, platelet activation and degranulation, blood coagulation and the acute phase response.
One of the first large-scale COVID-19 multiomic analyses to emerge was triggered by the foresight of Ariel Jaitovich, a pulmonary and critical care physician at the Albany Medical College in New York. Jaitovich saw first-hand how COVID-19 symptoms could range in severity and imagined a collaborative study that would provide relevant molecular insights. Expertise was subsequently pooled from the Coon Laboratory at the University of Wisconsin-Madison and the Morgridge Institute for Research to produce a large-scale multiomic analysis of COVID-19 severity.2
Together, the group performed RNA sequencing and high-resolution mass spectrometry (MS) on 128 plasma samples from people with and without COVID-19, and correlated leukocyte mRNA expression and plasma protein, metabolite and lipid levels with a range of clinical data and patient outcomes. “Our end goal was to use our resources and shared expertise to get the data to people as fast as we could, without agonizing over our own analysis,” says Ian Miller, research data scientist at the Coon Laboratory. “Data on COVID was coming out like a firehose, and it still is.”
Miller recalls how different therapeutic molecules could be observed among the metabolomics measurements, providing what he describes a “baked-in sanity check or control.” 219 biomolecules were highly correlated with COVID-19 status and severity, as were dense biomolecule clusters. Of note, one identified cluster contained lipids – plasmenyl–phosphatidylcholines and high-density lipoproteins – categories that are significantly associated with COVID-19 status and severity.
The characterization of COVID-19 pathophysiology in multiomic studies allows for the identification of potential therapeutic opportunities – several of which were postulated in the publication. 2 For example, the authors highlight the potential use of statins or other therapies aimed at restoring high-density lipoprotein (HDL). Reduced levels of circulating HDL in COVID-19 patients have been reported in other studies, potentially contributing to the stress and inflammation that aggravates COVID-19 pathophysiology.3 Ultimately, the hope is that multiomic data can also be used to support more accurate predictions of patient outcomes.
Obtaining the Whole Genome Sequence of SARS-CoV-2
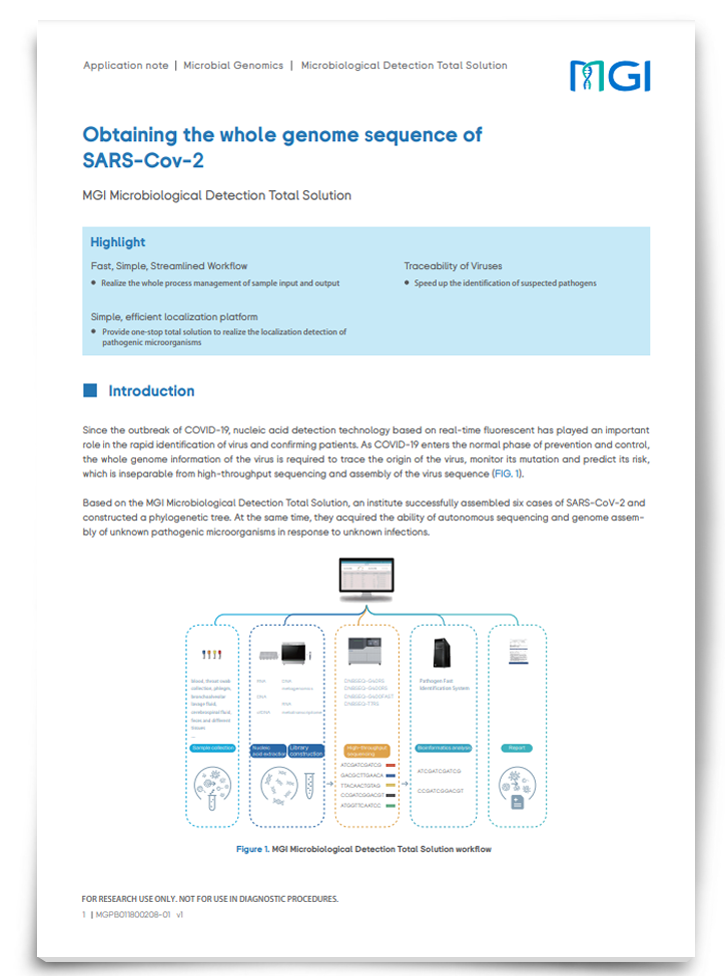
As COVID-19 enters the normal phase of prevention and control, whole genome information for SARS-CoV-2 is required to trace the origin of the virus, identify and monitor mutations, and predict its risk, which is inseparable from high-throughput sequencing and assembly of the virus sequence. Download this app note to discover a detection solution that provides a fast, simple and streamlined workflow and expedites the identification of suspected pathogens.
Download App NoteSponsored Content
Investigating cellular hallmarks of severe COVID-19
Studying the mechanisms of thrombotic complications in COVID-19 is a priority, given the high prevalence of blood clotting problems in critically ill cases.4 Several studies have pointed to altered platelet immune cell interactions and the presence of megakaryocytes in affected lungs.5,6 To investigate further, Bernardes and colleagues employed a longitudinal multiomics analysis to characterize the cellular features of severe COVID-19 cases.7 Analyzing the bulk transcriptome, bulk DNA methylome, and single-cell transcriptome of over 358,000 cells from peripheral blood enabled cellular changes to be characterized over time. Elevated levels of circulating megakaryocytes were identified in severe COVID-19 cases and linked to heightened inflammatory parameters, alongside changes in expression revealed by bulk RNA sequencing data. The multiomic approach provides greater context when studying dynamic changes of circulating cells in the blood. For example, genome-wide hypomethylation was observed at each time point compared with healthy controls, supporting earlier suggestions of epigenetic changes contributing to systemic inflammatory states.8
Making multiomic COVID-19 data more accessible
As summarized by Abadi, sophisticated tools are needed if multiomics data are to be harnessed: “The sequencing data generated by these assays require new data structures to streamline the downstream analyses. These data are also highly dimensional and gaining direct insight from them is beyond our limited intuition.
Tailored computational and statistical methods are pivotal in unmasking the hidden and often interesting patterns in the data, testing hypotheses generated by researchers, and/or creating new insights which could help generate new hypotheses.” As a result, many groups are developing tools designed to help bridge the gap between the generation and understanding of data. mixOmics is one such example – a free and open-source multiomics tool kit, and the result of an international, multi-university collaboration. Abadi has had a central role in the implementation and improvement of novel multivariate statistical methods for mixOmics, while also developing powerful visualization tools to present outcomes in a more compelling and intuitive way.
The need for data visualization tools is not lost on the Morgridge/University of Wisconsin-Madison collaborators. To shift data analysis from a “firehose” experience to something more manageable, the group transferred the data onto an interactive webtool – covid-omics.app –allowing access to the wider scientific community. “We want to share what we have produced – that’s our model. And that’s inherent to the funding mechanism for most of these projects too, a lot of this work is federally funded” says Katie Overmyer, associate director of the Laboratory for Biomolecular Mass Spectrometry at the Morgridge Institute for Research.
Transform Your Research With High-Purity Lipids

Whether you specialize in lipidomics, or lipids are a regular part of your research, you know the critical role they play and the inherent challenges they present. Download this brochure to discover high-purity, natural and synthetic phospholipids, sphingolipids, sterols, fluorescent lipids, adjuvants and lipid-binding antibodies. Find out how you can deliver high purity and yields in a single step.
View BrochureSponsored Content
Preparing for a data-driven world of biological research
By harnessing technologies such as machine learning, RNA sequencing and MS, omics data can be combined with clinical information to provide researchers with a wealth of data to explore. For Miller, data literacy is an essential tool he would recommend biologists add to their toolkits: “The more data literacy you can pick up and the more basic programming you can learn, the more it will allow you to understand and share your work.” As researchers work to disseminate and combat the highly variable clinical course of COVID-19 infection, the ability to share valuable data with a wider audience – while providing helpful tools for visualization and analysis – has never been so critical.
References
- Huang S, Chaudhary K, Garmire LX. More is better: recent progress in multi-omics data integration methods. Front Genet. 2017;8:84. doi: 10.3389/fgene.2017.00084.
- Overmyer KA, Shishkova E, Miller IJ, et al. Large-scale multi-omic analysis of COVID-19 severity. Cell Systems. 2021;12(1):23-40.e7. doi: 10.1016/j.cels.2020.10.003.
- Begue F, Tanaka S, Mouktadi Z, et al. Altered high-density lipoprotein composition and functions during severe COVID-19. Scientific Reports. 2021;11(1):2291. doi: 10.1038/s41598-021-81638-1.
- Deshpande C. Thromboembolic findings in COVID-19 autopsies: pulmonary thrombosis or embolism? Ann Intern Med. 2020;173(5):394-395. doi: 10.7326/M20-3255.
- Leppkes M, Knopf J, Naschberger E, et al. Vascular occlusion by neutrophil extracellular traps in COVID-19. EBioMedicine. 2020;58:102925. doi: 10.1016/j.ebiom.2020.102925.
- Meyerholz DK, McCray PB. Illuminating COVID-19 lung disease through autopsy studies. EBioMedicine. 2020;57:102865. doi: 10.1016/j.ebiom.2020.102865.
- Bernardes JP, Mishra N, Tran F, et al. Longitudinal multi-omics analyses identify responses of megakaryocytes, erythroid cells, and plasmablasts as hallmarks of severe COVID-19. Immunity. 2020;53(6):1296-1314.e9. doi: 10.1016/j.immuni.2020.11.017.
- Lorente-Sorolla C, Garcia-Gomez A, Català-Moll F, et al. Inflammatory cytokines and organ dysfunction associate with the aberrant DNA methylome of monocytes in sepsis. Genome Medicine. 2019;11(1):66. doi: 10.1186/s13073-019-0674-2.
Article updated September 6, 2021, to remove the hyphen from "Multi-omics".





