Cryo Electron Microscopy: Principle, Strengths, Limitations and Applications
Complete the form below to unlock access to ALL audio articles.
Cryo electron microscopy (cryo EM) has revolutionized our understanding of the intricate molecular machinery that governs life. This cutting-edge imaging technique allows scientists to peer into the microscopic world with unprecedented clarity and detail, revealing the hidden structures of proteins, viruses and cellular components. With its origins rooted in the pioneering work of scientists like Jacques Dubochet,1 Joachim Frank2 and Richard Henderson,3 cryo EM has evolved into a powerful tool that has garnered numerous accolades, including the 2017 Nobel Prize in Chemistry.4
What is cryo EM?
Cryo EM is an advanced imaging technique used to elucidate the three-dimensional (3D) structure of biological molecules and complexes at near-atomic resolution. In this technique, the sample is rapidly frozen to temperatures below -150 °C, trapping it in vitreous ice. The sample is then imaged from different angles using an electron microscope, where a beam of electrons passes through the specimen, resulting in a set of two-dimensional (2D) projections. Advanced computational algorithms are then used to reconstruct a 3D model of the sample from these projections.5, 6, 7
Cryo EM displays a series of important differences compared to convential EM that makes it a powerful tool for structural biology, providing insights into previously inaccessible areas of research. The main differences between the two techniques are summarised in Table 1.5, 6, 7
Table 1: Comparison of the main features of convential EM and cryo EM.
|
| Conventional EM | Cryo EM |
| Sample preparation | Involves fixation, staining, dehydration and the use of chemical compounds or polymers | Involves flash-freezing techniques (vitrification) |
| Sample integrity | Native structures can be altered and artifacts may appear | Native structures are well preserved |
| Resolution | High resolution (nm) | Higher resolution, near-atomic (Å) |
| Specimen types | Ideal for cellular and subcellular structures | Ideal for biomolecules and complexes |
| Amount of sample | Larger amounts of sample are required | Minimal amounts of biomolecules or complexes can be analyzed |
The development of cryo-EM (Figure 1) was a consequence of the limitations of conventional EM. Since the publication of the first 3D structure of a biological specimen obtained using EM in 1968,8 scientists tried to find strategies to circumvent the main problems of this technique, i.e., sample destruction caused by the high energy electron beam,9 alteration of the native structures and generation of undesiderable artifacts derived from the sample preparation process10 and low resolution when strong diffraction is present.
Several solutions were developed to assess this issues, including the use of heavy metal salts to embed the sample11 or the replacement of the water present in the sample by a glucose solution in order to reduce sample damage during the analysis.3 In this context, some scientists suggested that cooling down the samples to cryogenic temperatures would be expected to avoid water evaporation and reduce the damage caused by the electron radiation. However, this approach implies the formation of crystalline ice in the sample, which has two main undesiderable effects: ice crystals can induce damage in the sample structures and they also cause a strong electron diffraction that dramatically reduces resolution.12
The cryo EM era started in the early 1980s with the development of a methodology that applied a flash freezing approach that rapidly cools the samples at temperatures below the water glass-transition temperature (around -140 °C) within milliseconds. This procedure avoided the nucleation of crystals and generated amorphous ice in a process known as “vitrification”. 1
In the 1990s, a new strategy was established to improve cryo EM resolution by averaging the images from many copies of the same specimen.13 That milestone, together with more recent technological improvements, such as the development of direct electron detectors and better computational methods, have led to improved imaging capabilities that revolutionized the field.6 As a consequence, in the last decades, cryo EM has emerged as a technique able to compete with traditional structural techniques, such as X-ray crystallography or nuclear magnetic resonance (NMR) spectroscopy.

Figure 1: Timeline showing the historical development of cryo EM. Credit: Technology Networks.
How does cryo EM work?
The cryo EM workflow (Figure 2) begins with the preparation of a sample, typically a purified protein or complex in solution. The sample solution is placed onto a specialized grid and then it is rapidly frozen by plunging it into a cryogenic fluid, such as liquid ethane. This transforms the water into amorphous ice, causing the vitrification of the sample and preserving the native structure of the proteins. The vitrification procedure can be performed following different methodologies, some of which are optimized to capture short-lived molecular states.14, 15
Next, the vitrified sample is placed on the grid and transferred to the cryo EM microscope where it is maintained at extremely low temperatures to preserve sample integrity. The microscope is calibrated and aligned, and data acquisition begins. An electron beam is directed at the sample, producing a series of 2D projection images from different angles. These 2D images are processed to correct for artefacts and improve the signal-to-noise ratio. Individual particles within the images are identified through particle picking, and a large dataset of particles is collected. Computational algorithms are then employed to reconstruct a 3D density map from the particle images. The resulting 3D density map provides a representation of the electron scattering intensities of the sample, allowing scientists to gain insights into its structure. Atomic models are fitted into the density map and refined iteratively to represent the experimental data accurately. The refined models are validated and analyzed to understand the structural features of the studied molecules. 5, 14, 15, 16, 17

Figure 2: Typical cryo EM workflow. Credit: Hiramano92, reproduced under the Creative Commons Attribution 4.0 International (CC BY 4.0) license.
The vitrification of the sample using cryogenic fluids, the most definitory feature of cryo EM, can be applied to both traditional variants of conventional EM: transmission electron microscopy (TEM) and scanning electron microscopy (SEM). This means that cryo EM can be divided into two main types (Figure 3), a comparison of which is summarized in Table 2:
- Cryo TEM: in this technique, the vitrified sample is loaded into a high-vacuum TEM instrument. Inside the TEM, an electron beam is generated and directed onto the sample. As the beam passes through the sample, it interacts with the electrons, resulting in scattering and absorption. This interaction leads to the formation of a projection image on a detector, capturing the 2D structural information of the sample. To obtain a 3D structure, multiple projection images are collected by tilting the sample at different angles within the microscope. These images are then processed using advanced computational algorithms to generate a 3D density map of the sample. This density map represents the distribution of electrons within the sample and provides insights into the atomic arrangement of the molecules.18
- Cryo SEM: this technique is used to study the surface structure and topography of the vitrified samples. The vitrified sample is usually fractured or cleaved to expose its internal surfaces and then transferred to a SEM instrument. This microscope generates a focused electron beam that scans the sample's surface. As the beam interacts with the sample, secondary and backscattered electrons are emitted, which are then detected and used to create an image of the sample's surface.19

Figure 3: Examples of images obtained with cryo TEM (left) and cryo SEM (right) of Salmonella Senftenberg (upper row) and vaccinia virus (lower row). Credit: Adapted from Golding et al., 2016,20 reproduced under the Creative Commons Attribution 4.0 International (CC BY 4.0) license.
Table 2: Comparison of the main features of cryo TEM and cryo SEM.
|
| Cryo TEM | Cryo SEM |
| Equipment | Transmission electron microscope | Scanning electron microscope |
| Electron beam | It is transmitted through the sample | It is focused on the surface |
| Sample thickness | Thin samples | Any thickness |
| Information obtained | Internal structure of the sample | Surface topography and morphology of the sample |
| Resolution | High resolution (near-atomic) | Lower resolution |
| Applications | Molecular structures and interactions Suitable for single-particle analysis | Surface characterization Suitable for large-scale structures or materials |
Cryo EM vs cryo ET
Cryo electron tomography (cryo ET) is a technique that combines the principles of cryo EM and tomography, and it is especially useful to investigate cellular and subcellular 3D structures. This approach is based on the acquisition of a series of images taken at different tilt angles of the specimen with respect to the electron beam. The different planes collected are combined using computational algorithms to reconstruct the 3D structure. 7, 15, 21, 22
The typical workflow of cryo ET is similar to that of cryo EM (Figure 4), including the preparation of the biological sample by isolation and preservation through vitrification. The sample is placed onto a grid, forming a thin, even layer, and then the grid is transferred to a cryo-TEM microscope to acquire images of the sample from various angles by tilting the grid. The images captured constitute a tomographic dataset, which contains a series of 2D projections of the sample that provide information about its structure from different orientations. These 2D projections are aligned and corrected to minimize any misalignment or distortion, and finally, iterative algorithms or weighted back-projection is employed to reconstruct a 3D tomogram. This tomogram is analyzed to gain insight about the sample structure and organization. Sometimes, a subtomogram averaging is performed in order to improve the signal-to-noise ratio and extract information at higher resolution of specific regions of the tomogram. 21, 23, 24, 25, 26

Figure 4: A typical cryo ET workflow. Credit: Technology Networks.
The advantages of cryo ET include its ability to study intact cells and tissues, preserving the spatial relationships between different cellular components. It provides 3D information at nanometric resolution and it is also suitable for studying dynamic processes within cells, as it allows for time-resolved imaging that capture structural changes over time. On the other hand, this technique also has some limitations, most of them also typical of cryo EM, such as the complex sample preparation or the necessity of using low dose electron beams to avoid sample damage, which results in lower signal-to-noise ratio. Additionally, the sample thickness is limited to ~ 500 nm and the range of tilt angle is limited so the information obtained is partial. 7, 21, 25, 26
All these features make cryo ET more suitable for the study of cellular structures within their native environment, whereas cryo EM provides high-resolution imaging of individual macromolecular complexes, providing atomic-level details. Both techniques are often complementary in addressing different biological questions, ranging from molecular structures to cellular organization and dynamics.
Strengths and limitations of cryo EM
Cryo EM has emerged as a powerful technique in structural biology, offering several strengths and limitations: 6, 14, 15, 16, 17
- High, near-atomic resolution imaging.
- Sample preservation enables the study of samples in their near-native state.
- It can be applied to a wide range of biological samples, including proteins, viruses, cells and tissues.
- It can be applied to the study of large, flexible complexes and assemblies that are challenging for other techniques.
- It enables single particle analysis with no size limit or need for crystallization.
Limitations
- It requires certain homogeneity in the particle population to achieve high-resolution determination.
- It generates large datasets that are computationally demanding.
- There are resolution limitations and sometimes structural details cannot be well-resolved.
- Larger samples are difficult to study.
- Cryo EM equipment is costly and requires expert users.
When compared to other structural biology techniques, such as NMR spectroscopy and X-ray crystallography, cryo EM has distinct advantages and limitations. NMR spectroscopy is well-suited for studying the dynamics and interactions of smaller proteins in solution, providing information about atomic-level structures and dynamics. However, NMR faces limitations in resolving larger protein complexes and membrane proteins. Cryo EM, with its ability to study larger and more complex structures, complements NMR by providing high-resolution structures of such macromolecules. 6, 14, 27
X-ray crystallography, on the other hand, has been a prominent technique for determining atomic-level structures of proteins. It is particularly effective for small to medium-sized proteins and well-ordered crystalline samples. However, some proteins are challenging to crystallize, and their structures can be distorted during the crystallization process. Cryo EM offers an alternative for obtaining structures without the need for crystal formation, making it more applicable to a broader range of proteins and complexes. 6, 14, 28
In summary, cryo EM is best suited for studying large and flexible macromolecules, complexes with conformational heterogeneity and membrane proteins. Its ability to visualize samples in their native state and its compatibility with computational analysis contribute to its strength. While cryo EM complements NMR spectroscopy and X-ray crystallography by addressing their limitations, each technique has its unique strengths, making them valuable tools in the structural biology toolbox. The choice of technique depends on the specific research question and the characteristics of the biomolecule being investigated. 6, 14
Applications of cryo EM
Cryo EM finds applications in various fields of scientific research, enabling insights into the structural organization and dynamics of biological macromolecules. Here are some examples of applications of cryo EM.
- Structural biology: Cryo EM has revolutionized structural biology by determining high-resolution structures of challenging targets such as membrane proteins, large macromolecular assemblies and flexible or heterogeneous complexes that were inaccessible to traditional structural biology techniques (Figure 5). This has facilitated the understanding of protein structures, functions, mechanisms and interactions.

Figure 5: Cryo EM structure of the light harvesting complex 2 from Marichromatium purpuratum obtained with a resolution of 2.4 Å. Credit: Adapted from Gardiner et al., 2021),29 reproduced under the Creative Commons Attribution 4.0 International (CC BY 4.0) license. - Virology: Cryo EM allows the visualization of viral capsids, envelopes and surface proteins, aiding in vaccine development, antiviral drug design and understanding viral assembly, replication and infection mechanisms. One of the most recent advances in this field is related to the structure of the spike protein of the SARS-CoV2 omicron virus (Figure 6).

Figure 6: Cryo EM structure of the full-length SARS-CoV2 omicron spike protein. Credit: Adapted from Ye et al., 2022,30 reproduced under the Creative Commons Attribution 4.0 International (CC BY 4.0) license. - Cell biology: Cryo EM provides insights into cellular architecture and organization by visualizing cellular organelles, cytoskeletal networks and molecular complexes within cells. It helps unravel fundamental cellular processes like mitosis, endocytosis, intracellular transport and organelle biogenesis (Figure 7).

Figure 7: Cryo ET applied to the study of the mitotic spindle. A: Tomographic slice showing a cell during metaphase (ch: chromosomes; p: spindle poles). B: 3D reconstruction of the same spindle shown in A, stacking the corresponding serial tomograms. Credit: Adapted from Kiewisz et al., 2021,31 reproduced under the Creative Commons Attribution 4.0 International (CC BY 4.0) license. - Drug discovery: Cryo EM plays a vital role in drug discovery and development by elucidating the structures of drug targets, such as G protein-coupled receptors (GPCRs) and ion channels (Figure 8). The technique aids in rational drug design, understanding ligand binding mechanisms and optimizing drug efficacy and specificity.
 Figure 8: Cryo EM structure of the cyclic nucleotide-gated ion channel SthK. (PDB: 6CJQ). Credit: Nimigean CM, Rheinberge, J, reproduced under the Creative Commons 0 1.0 Universal (CC0 1.0) Public Domain Dedication.
Figure 8: Cryo EM structure of the cyclic nucleotide-gated ion channel SthK. (PDB: 6CJQ). Credit: Nimigean CM, Rheinberge, J, reproduced under the Creative Commons 0 1.0 Universal (CC0 1.0) Public Domain Dedication. - Neurobiology: Cryo EM is employed to study the structures of neuronal synapses, neurotransmitter receptors and macromolecular complexes associated with neurodegenerative diseases (Figure 9). It contributes to unraveling the molecular mechanisms underlying synaptic transmission, neuronal signaling and neurological disorders.

Figure 9: Cryo ET of synaptic vesicles in primary hippocampal neurons. A: A tomographic slice of the neuron synapse. B: 3D reconstruction of the tomogram showing the distribution of synaptical vesicles (SV). PostMito = postsynaptic mitochondria; PrM = presynaptic membrane; PoM = postsynaptic membrane; DCV = dense core vesicles; EV = endosomal vesicles; MT = microtubules; Mito = mitochondria. Credit: Adapted from Bäuerlein et al., 2017),32 reproduced under the Creative Commons Attribution 4.0 International (CC BY 4.0) license. - Biochemistry and molecular biology: Cryo EM assists in studying macromolecular assemblies involved in crucial cellular processes, including DNA replication, transcription, translation and protein folding. It provides valuable structural information for understanding the molecular mechanisms underlying these biological processes (Figure 10).

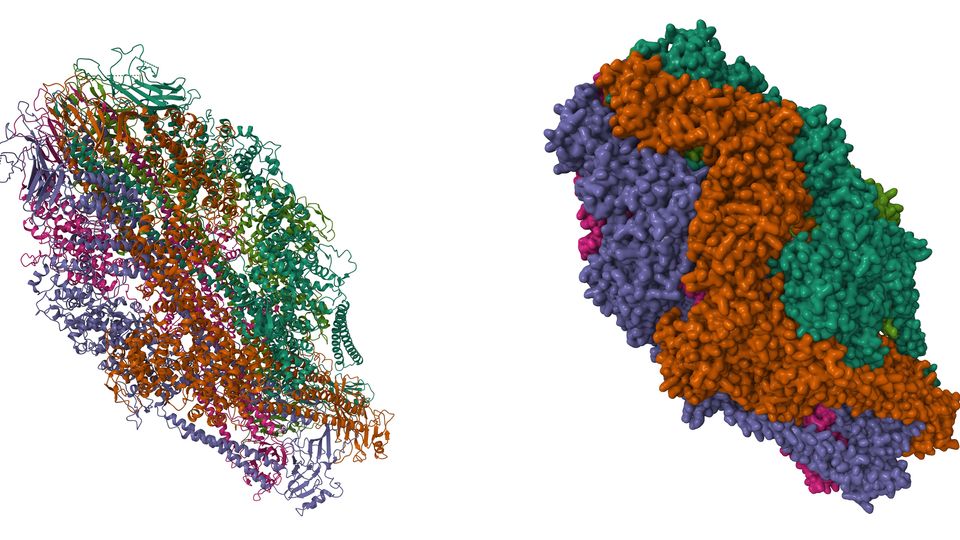
Figure 10: Cryo EM structure of the human RNA polymerase III in the apo form. (PDB: 7A6H). Credit: Girbig M, Misiaszek AD, Vorlaender MK, Mueller CW, reproduced under the Creative Commons 0 1.0 Universal (CC0 1.0) Public Domain Dedication.
In conclusion, cryo EM has emerged as a transformative technique in the field of structural biology, revolutionizing our understanding of complex macromolecular assemblies and cellular architectures. By enabling the visualization of biological samples in their near-native state and at high resolution, cryo EM has provided unprecedented insights into the structures and functions of diverse biomolecular systems. Its versatility allows researchers to study a wide range of samples, from individual proteins and viruses to intact cells and tissues. Cryo EM's unique ability to visualize large and flexible complexes, often challenging for other techniques, has led to groundbreaking discoveries in fields such as drug design and vaccine development. However, cryo-EM shows some limitations, related to sample heterogeneity and the need for significant computational resources. Nonetheless, continuous advancements in hardware and software, coupled with hybrid approaches that combine cryo EM with other structural biology techniques, continue to expand the scope and impact of this powerful method.
1. Dubochet J and McDowall AW. Vitrification of pure water for electron microscopy. J Microscopy. 1981; 124(3):3-4. doi:10.1111/j.1365-2818.1981.tb02483.x
2. Frank J, Radermacher M, Penczek P, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996; 116(1):190-199. doi:10.1006/jsbi.1996.0030
3. Unwin PNT and Henderson R. Molecular structure determination by electron microscopy of unstained crystalline specimens. J Mol Biol. 1975; 94(3):425-432. doi:10.1016/0022-2836(75)90212-0
4. The Nobel Prize in Chemistry 2017. Press release. The Nobel Prize. https://www.nobelprize.org/uploads/2018/06/press-41.pdf. Published 2017. Accessed June 25, 2023.
5. Bai X-C, McMullan G and Scheres SHW. How cryo-EM is revolutionizing structural biology. Trends Biochem Sci. 2014; 40(1):49-57. doi:10.1016/j.tibs.2014.10.005
6. Benjin X and Ling L. Developments, applications, and prospects of cryo-electron microscopy. Protein Sci. 2019; 29:872-882. doi:10.1002/pro.3805
7. Milne JLS, Borgnia MJ, Bartesaghi A et al. Cryo-electron microscopy. A primer for the non-microscopist. FEBS J. 2013; 280(1):28-45. doi:10.1111/febs.12078
8. De Rosier DJ and Klug A. Reconstruction of three dimensional structures from electron micrographs. Nature. 1968; 217(5124):130-134. doi:10.1038/217130a0
9. Cheng Y, Glaeser RM and Nogales E. How cryo-EM became so hot. Cell. 2017; 171(6):1229-1231. doi:10.1016/j.cell.2017.11.016
10. Dubochet J. Cryo-EM. The first thirty years. J Microsc. 2012; 245(3):221-224. doi:10.1111/j.1365-2818.2011.03569.x
11. Brenner S and Horne RW. A negative staining method for high resolution electron microscopy of viruses. Biochim Biophys Acta. 1959; 34:103-110. doi:10.1016/0006-3002(59)90237-9
12. Fernández-Morán H. Low-temperature preparation techniques for electron microscopy of biological specimens based on rapid freezing with liquid helium II. Ann NY Acad Sci. 1960; 13(85):689-713. doi:10.1111/j.1749-6632.1960.tb49990.x
13. Henderson R, Baldwin JM, Ceska TA, Zemlin F, Beckmann E and Downing KH. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990; 213(4):899-929. doi:10.1016/S0022-2836(05)80271-2
14. Frank J. Single-particle imaging of macromolecules by cryo-electron microscopy. Annu Rev Biophys Struct. 2002; 31:303-319. doi:10.1146/annurev.biophys.31.082901.134202
15. Guaita M, Watters SC and Loerch S. Recent advances and current trends in cryo-electron microscopy. Curr Op Struct Biol. 2022; 77:102484. doi:10.1016/j.sbi.2022.102484
16. Earl LA, Falconieri V, Milne JLS and Subramaniam S. Cryo-EM: beyond the microscope. Curr Op Struct Biol. 2017; 46:71-78. doi:10.1016/j.sbi.2017.06.002
17. Nogales E and Scheres SHW. Cryo-EM: A unique tool for the visualization of macromolecular complexity. Mol Cell. 58(4):677-689. doi:10.1016/j.molcel.2015.02.019
18. Harris JR. Transmission electron microscopy in molecular structural biology: A historical survey. Arch Biochem Biophys. 2015; 581:3-18. doi:10.1016/j.abb.2014.11.011
19. Rigort A and Plitzko JM. Cryo-focused-ion-beam applications in structural biology. Arch Biochem Biophys. 2015; 581:122-130. doi:10.1016/j.abb.2015.02.009
20. Golding CG, Lamboo LL, Beniac DR and Booth TF. The scanning electron microscope in microbiology and diagnosis of infectious disease. Sci Rep. 2016; 6:26516. doi:10.1038/srep26516
21. Lučić V, Rigort A and Baumeister W. Cryo-electron tomography: The challenge of doing structural biology in situ. J Cell Biol. 2013; 202(3):407-419. doi:10.1083/jcb.201304193
22. Koning RI, Koster AJ and Sharp TH. Advances in cryo-electron tomography for biology and biomedicine. Ann Anat. 2018; 217:82-96. doi:10.1016/j.aanat.2018.02.004
23. Koning RI and Koster AJ. Cryo-electron tomography in biology and medicine. Ann Anat. 2009; 191:427-445. doi:10.1016/j.aanat.2009.04.003
24. Turk M and Baumeister W. The promise and the challenges of cryo-electron tomography. FEBS Lett. 2020; 594(20):3243-3261. doi:10.1002/1873-3468.13948
25. Hylton RK and Swulius MT. Challenges and triumphs in cryo-electron tomography. iScience. 2021; 24(9):102959. doi:10.1016/j.isci.2021.102959
26. Baumeister W. Cryo-electron tomography: A long journey to the inner space of cells. Cell. 2022; 185(15):2649-2652. doi:10.1016/j.cell.2022.06.034
27. Markwick PRL, Malliavin T and Nilges M. Structural biology by NMR: Structure, dynamics and interactions. PLoS Comput Biol. 2008; 4(9):e1000168. doi:10.1371/journal.pcbi.1000168
28. Ameh ES. A review of basic crystallography and X-ray diffraction applications. Int J Adv Manuf Technol. 2019; 105:3289-3302. doi:10.1007/s00170-019-04508-1
29. Gardiner AT, Naydenova K, Castro-Hartmann P, Nguyen-Phan TC, Russo CJ, Sader K et al. The 2.4 Å cryo-EM structure of a heptameric light-harvesting 2 complex reveals two carotenoid energy transfer pathways. Sci Adv. 2021; 7(7):eabe4650. doi:10.1126/sciadv.abe4650
30. Ye G, Liu B and Li F. Cryo-EM structure of a SARS-CoV-2 omicron spike protein ectodomain. Nat Comm. 2022; 13:1214. doi:10.1038/s41467-022-28882-9
31. Kiewisz R, Fabig G, Conway W, Baum D, Needleman D and Müller-Reichert T. Three-dimensional structure of kinetochore-fibers in human mitotic spindles. eLife. 2022; 11:e75459. doi:10.7554/eLife.75459
32. Radhakrishnan A, Li X, Grushin K and Rothman JE. Symmetrical arrangement of proteins under release-ready vesicles in presynaptic terminals. Proc Natl Acad Sci U.S.A. 2021; 118(5):e2024029118. doi:10.1073/pnas.2024029118






